Research Projects
The Spine Biomechanics group comprising engineers from ETH Zurich and surgeons from the Balgrist University Hospital. Together they conduct translational research at the interface between fundamental biomechanical research and the development of clinically applicable methods. The interdisciplinary approach guarantees a high level of expertise required to address relevant clinical issues. The group has established a fully equipped spine lab with custom test setups for biomechanical in-vitro experiments that is on par with the worldwide leading biomechanical spine laboratories. This equipment has been used for a multitude of biomechanical studies on human spine cadavers to investigate and characterize human anatomy and tissue properties. The obtained results and insights are translated into patient-specific, biomechanical models that are used to optimize pre-operative surgical planning.
Automated Extraction of Spinopelvic Parameters from EOS X-Ray Images
This project aims to create a fully automated deep-learning pipeline that extracts spinopelvic parameters from biplanar x-ray images.
In a population with an increasing rate of spinal pathologies, the frontal and sagittal spinopelvic alignment is gaining more importance for the evaluation and treatment of such spinal disorders. However, manually gathering position data from spinal x-ray images is a tedious and time-consuming process. Different approaches have been introduced in the last years to automatically determine spinopelvic parameters. Recent research focused either on the frontal or sagittal spine, or evaluated only parts of the spine, e.g., the lumbopelvic or cervical region. Other approaches were limited to young patients. To our knowledge, no approach has yet been made to create a fully automated pipeline for gathering both complete frontal and spinal alignment information from EOS images in patients of all ages.
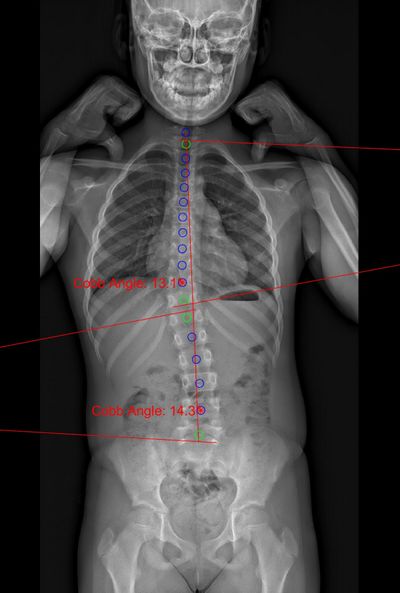
Figure 1: Cobb angle (frontal).
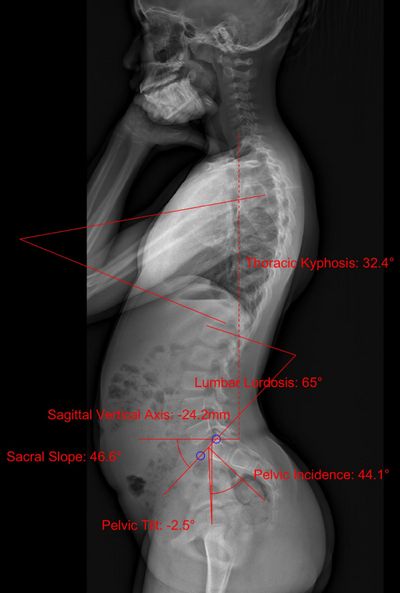
Figure 2: Lumbar lordosis, thoracic kyphosis, sacral slope, pelvic tilt, pelvic incidence, sagittal vertical axis (lateral).
The fully automated pipeline can identify positional data of the pelvis and vertebrae as well as the following spinopelvic parameters: cobb angle, lumbar lordosis, thoracic kyphosis, sacral slope, pelvic tilt, pelvic incidence, sagittal vertical axis.
The workflow from a raw EOS image to the prediction of spinopelvic parameters is divided into three steps:
- pre-processing the image data in Matlab,
- segmentation of the regions of interest based on a deep-convolutional U-net implemented in TensorFlow (Python),
- post-processing of segmented areas and computation of spinopelvic parameters in Matlab.
The automatic approach allows a quantitative assessment of large data sets and might help to identify patterns in clinical presentations or even supports the prediction of outcomes after surgical interventions.



